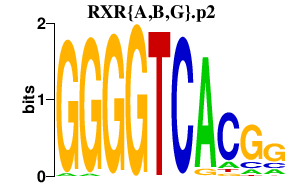
|
chr12_-_112910475
|
24.320
|
NM_021273
|
Ckb
|
creatine kinase, brain
|
|
chr17_+_43938789
|
23.304
|
NM_207649
|
Rcan2
|
regulator of calcineurin 2
|
|
chr11_+_101329457
|
22.434
|
|
Rnd2
|
Rho family GTPase 2
|
|
chr10_+_59569052
|
20.095
|
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2
|
|
chr16_-_85173764
|
19.837
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chr9_+_110148568
|
19.276
|
|
Cspg5
|
chondroitin sulfate proteoglycan 5
|
|
chr13_-_25029458
|
18.788
|
NM_172532
|
Aldh5a1
|
aldhehyde dehydrogenase family 5, subfamily A1
|
|
chr16_-_85173930
|
18.750
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chr16_-_85173851
|
18.743
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chr16_-_85173918
|
18.587
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chr16_-_85173978
|
17.790
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chr8_+_86496844
|
17.381
|
NM_008854
|
Prkaca
|
protein kinase, cAMP dependent, catalytic, alpha
|
|
chr11_+_78139476
|
16.962
|
|
Aldoc
|
aldolase C, fructose-bisphosphate
|
|
chr2_-_25325248
|
16.828
|
NM_008963
|
Ptgds
|
prostaglandin D2 synthase (brain)
|
|
chr9_-_31019412
|
16.425
|
|
Aplp2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr10_+_59569206
|
16.354
|
|
Spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2
|
|
chr16_-_18629938
|
15.648
|
NM_213614
|
Sept5
|
septin 5
|
|
chr2_-_25325196
|
15.512
|
|
Ptgds
|
prostaglandin D2 synthase (brain)
|
|
chr16_-_84955488
|
15.177
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chr7_-_52124950
|
13.938
|
|
Ptov1
|
prostate tumor over expressed gene 1
|
|
chr2_-_148558110
|
13.847
|
NM_019632
|
Napb
|
N-ethylmaleimide sensitive fusion protein attachment protein beta
|
|
chr7_-_52125157
|
13.687
|
NM_133949
|
Ptov1
|
prostate tumor over expressed gene 1
|
|
chr11_+_78136681
|
13.470
|
|
Aldoc
|
aldolase C, fructose-bisphosphate
|
|
chr11_+_101329615
|
13.358
|
NM_009708
|
Rnd2
|
Rho family GTPase 2
|
|
chr13_-_78338131
|
13.009
|
NM_010151
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr17_+_43938905
|
12.929
|
|
Rcan2
|
regulator of calcineurin 2
|
|
chr15_-_83936183
|
12.905
|
NM_013873
|
Sult4a1
|
sulfotransferase family 4A, member 1
|
|
chr7_-_149276174
|
12.595
|
NM_008748
|
Dusp8
|
dual specificity phosphatase 8
|
|
chr4_-_151235861
|
12.526
|
NM_001081557
NM_001195565
|
Camta1
|
calmodulin binding transcription activator 1
|
|
chr11_-_115158717
|
12.466
|
NM_178035
|
Fads6
|
fatty acid desaturase domain family, member 6
|
|
chr2_+_164793487
|
12.328
|
NM_020333
|
Slc12a5
|
solute carrier family 12, member 5
|
|
chr9_-_31019234
|
11.968
|
|
Aplp2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr7_-_52622289
|
11.693
|
|
Ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3
|
|
chr9_-_108363229
|
11.476
|
|
Klhdc8b
|
kelch domain containing 8B
|
|
chr9_-_103132606
|
11.426
|
NM_133977
|
Trf
|
transferrin
|
|
chr8_-_48374715
|
11.420
|
|
Stox2
|
storkhead box 2
|
|
chr8_-_73132125
|
11.294
|
NM_133772
|
Ssbp4
|
single stranded DNA binding protein 4
|
|
chr16_-_85173944
|
11.098
|
NM_001198823
NM_001198824
NM_001198825
NM_001198826
NM_007471
|
App
|
amyloid beta (A4) precursor protein
|
|
chr9_-_103132523
|
11.040
|
|
Trf
|
transferrin
|
|
chr2_+_177876858
|
10.982
|
|
Phactr3
|
phosphatase and actin regulator 3
|
|
chr2_+_164793698
|
10.949
|
|
Slc12a5
|
solute carrier family 12, member 5
|
|
chr18_-_16967754
|
10.790
|
|
Cdh2
|
cadherin 2
|
|
chr7_-_52622373
|
10.761
|
NM_029741
|
Ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3
|
|
chr14_+_64225366
|
10.402
|
NM_173393
|
Xkr6
|
X Kell blood group precursor related family member 6 homolog
|
|
chr9_+_75161942
|
10.108
|
NM_138719
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5
|
|
chr19_-_61302538
|
10.062
|
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage)
|
|
chr11_+_29593657
|
10.055
|
NM_194051
|
Rtn4
|
reticulon 4
|
|
chr11_+_78136690
|
9.942
|
|
Aldoc
|
aldolase C, fructose-bisphosphate
|
|
chr7_+_31092787
|
9.776
|
|
Clip3
|
CAP-GLY domain containing linker protein 3
|
|
chr13_-_78337844
|
9.719
|
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr2_+_177876710
|
9.686
|
|
Phactr3
|
phosphatase and actin regulator 3
|
|
chr11_-_69418965
|
9.644
|
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr5_-_118694869
|
9.594
|
NM_172998
NM_001109902
|
Rnft2
|
ring finger protein, transmembrane 2
|
|
chr7_-_129245303
|
9.468
|
NM_028177
|
Ndufab1
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1
|
|
chr5_+_18732863
|
9.366
|
NM_001170746
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr15_+_76490995
|
9.351
|
NM_010630
|
Kifc2
|
kinesin family member C2
|
|
chr8_-_73011217
|
9.289
|
NM_182991
|
Tmem59l
|
transmembrane protein 59-like
|
|
chr15_+_78744576
|
9.209
|
|
Pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase
|
|
chr2_+_106533281
|
9.167
|
|
Mpped2
|
metallophosphoesterase domain containing 2
|
|
chr5_+_37633317
|
9.130
|
NM_001136058
|
Crmp1
|
collapsin response mediator protein 1
|
|
chr2_+_135885126
|
9.110
|
|
6330527O06Rik
|
RIKEN cDNA 6330527O06 gene
|
|
chr15_+_103333440
|
9.045
|
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent
|
|
chr7_+_19661811
|
9.010
|
|
Dmwd
|
dystrophia myotonica-containing WD repeat motif
|
|
chr15_+_78744802
|
8.947
|
|
Pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase
|
|
chr5_+_108797254
|
8.918
|
|
Gm10419
|
predicted gene 10419
|
|
chr7_+_53693190
|
8.860
|
|
Kcnc1
|
potassium voltage gated channel, Shaw-related subfamily, member 1
|
|
chr5_-_31504348
|
8.846
|
NM_001177901
|
Zfp513
|
zinc finger protein 513
|
|
chrX_-_98615096
|
8.811
|
NM_001177985
NM_001177986
|
Zmym3
|
zinc finger, MYM-type 3
|
|
chr15_+_78258993
|
8.807
|
NM_001081367
|
Kctd17
|
potassium channel tetramerisation domain containing 17
|
|
chr10_+_90292453
|
8.782
|
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr1_-_25886551
|
8.748
|
NM_175642
|
Bai3
|
brain-specific angiogenesis inhibitor 3
|
|
chr17_+_25707771
|
8.748
|
|
2810468N07Rik
|
RIKEN cDNA 2810468N07 gene
|
|
chr8_-_112692071
|
8.727
|
NM_007586
|
Calb2
|
calbindin 2
|
|
chr11_+_71564204
|
8.705
|
NM_177618
|
Wscd1
|
WSC domain containing 1
|
|
chr18_-_16967557
|
8.666
|
NM_007664
|
Cdh2
|
cadherin 2
|
|
chr16_+_41533317
|
8.615
|
NM_175548
|
Lsamp
|
limbic system-associated membrane protein
|
|
chr15_-_83936038
|
8.548
|
|
Sult4a1
|
sulfotransferase family 4A, member 1
|
|
chr5_+_137729501
|
8.448
|
NM_009599
|
Ache
|
acetylcholinesterase
|
|
chr9_+_110147867
|
8.302
|
|
Cspg5
|
chondroitin sulfate proteoglycan 5
|
|
chr15_+_78744328
|
8.299
|
NM_020271
|
Pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase
|
|
chr8_-_48374755
|
8.182
|
NM_001114311
|
Stox2
|
storkhead box 2
|
|
chr15_-_98638230
|
8.150
|
|
Ddn
|
dendrin
|
|
chr2_+_28061156
|
8.140
|
NM_001038612
NM_019498
|
Olfm1
|
olfactomedin 1
|
|
chr17_-_24826812
|
8.135
|
|
Syngr3
|
synaptogyrin 3
|
|
chr1_+_78306890
|
8.068
|
NM_001004173
|
Sgpp2
|
sphingosine-1-phosphate phosphotase 2
|
|
chr3_-_89049756
|
8.011
|
NM_183037
|
Trim46
|
tripartite motif-containing 46
|
|
chr3_+_133903056
|
7.988
|
NM_001004367
|
Cxxc4
|
CXXC finger 4
|
|
chr11_+_69831951
|
7.978
|
NM_001109752
NM_007864
|
Dlg4
|
discs, large homolog 4 (Drosophila)
|
|
chr16_+_5885447
|
7.960
|
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr13_-_58009599
|
7.892
|
|
Spock1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 1
|
|
chr11_-_119409041
|
7.882
|
NM_008730
|
Nptx1
|
neuronal pentraxin 1
|
|
chr10_+_90292388
|
7.874
|
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr9_+_60560923
|
7.820
|
NM_026235
|
Larp6
|
La ribonucleoprotein domain family, member 6
|
|
chr5_+_37259906
|
7.784
|
|
Ppp2r2c
|
protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), gamma isoform
|
|
chr18_-_20904585
|
7.775
|
NM_019737
|
B4galt6
|
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 6
|
|
chr10_-_127058163
|
7.740
|
|
Lrp1
|
low density lipoprotein receptor-related protein 1
|
|
chr3_+_68298129
|
7.631
|
|
Schip1
|
schwannomin interacting protein 1
|
|
chrX_-_149471740
|
7.631
|
NM_023788
|
Mageh1
|
melanoma antigen, family H, 1
|
|
chr9_-_31019392
|
7.622
|
NM_001102455
NM_001102456
NM_009691
|
Aplp2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr6_-_4697009
|
7.611
|
NM_001130188
NM_001130189
NM_001130190
NM_001130191
NM_011360
|
Sgce
|
sarcoglycan, epsilon
|
|
chr17_-_46169322
|
7.605
|
NM_001025250
NM_001025257
NM_009505
|
Vegfa
|
vascular endothelial growth factor A
|
|
chr9_+_75162059
|
7.603
|
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5
|
|
chr4_+_137280366
|
7.602
|
|
Rap1gap
|
Rap1 GTPase-activating protein
|
|
chr14_-_55531726
|
7.597
|
NM_021551
|
Slc22a17
|
solute carrier family 22 (organic cation transporter), member 17
|
|
chr11_+_71564481
|
7.567
|
|
Wscd1
|
WSC domain containing 1
|
|
chr7_-_52622167
|
7.526
|
|
Ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3
|
|
chr10_+_79167197
|
7.523
|
|
Bsg
|
basigin
|
|
chr11_+_103510822
|
7.460
|
NM_001033212
|
Rprml
|
reprimo-like
|
|
chr4_-_109149038
|
7.452
|
NM_013876
|
Rnf11
|
ring finger protein 11
|
|
chr10_+_79167255
|
7.389
|
|
Bsg
|
basigin
|
|
chr14_+_67530044
|
7.379
|
NM_175498
|
Pnma2
|
paraneoplastic antigen MA2
|
|
chr16_+_18777032
|
7.343
|
|
Cldn5
|
claudin 5
|
|
chr9_-_56483379
|
7.181
|
|
Lingo1
|
leucine rich repeat and Ig domain containing 1
|
|
chr11_-_119903061
|
7.143
|
NM_001198787
|
Aatk
|
apoptosis-associated tyrosine kinase
|
|
chr11_+_92960603
|
7.107
|
NM_028296
|
Car10
|
carbonic anhydrase 10
|
|
chr4_+_124336307
|
7.053
|
|
Pou3f1
|
POU domain, class 3, transcription factor 1
|
|
chr7_+_19868009
|
7.050
|
NM_013648
|
Rtn2
|
reticulon 2 (Z-band associated protein)
|
|
chr2_+_106533615
|
7.030
|
NM_001143683
|
Mpped2
|
metallophosphoesterase domain containing 2
|
|
chr11_+_103510847
|
6.946
|
|
Rprml
|
reprimo-like
|
|
chr2_+_28368766
|
6.918
|
|
Ralgds
|
ral guanine nucleotide dissociation stimulator
|
|
chr16_+_18776955
|
6.906
|
|
Cldn5
|
claudin 5
|
|
chr6_+_113433408
|
6.874
|
|
Creld1
|
cysteine-rich with EGF-like domains 1
|
|
chr12_-_4848240
|
6.827
|
NM_016863
|
Fkbp1b
|
FK506 binding protein 1b
|
|
chr15_-_98638348
|
6.789
|
NM_001013741
|
Ddn
|
dendrin
|
|
chr5_+_34339258
|
6.751
|
|
Nat8l
|
N-acetyltransferase 8-like
|
|
chr11_-_3995299
|
6.727
|
NM_026443
|
Mtfp1
|
mitochondrial fission process 1
|
|
chr17_+_25854116
|
6.726
|
NM_022422
|
Gng13
|
guanine nucleotide binding protein (G protein), gamma 13
|
|
chr10_+_79167102
|
6.687
|
NM_001077184
NM_009768
|
Bsg
|
basigin
|
|
chr1_+_137043277
|
6.676
|
NM_026823
|
Arl8a
|
ADP-ribosylation factor-like 8A
|
|
chr8_-_4259126
|
6.670
|
NM_183315
|
Ctxn1
|
cortexin 1
|
|
chr11_-_116515361
|
6.630
|
|
Cygb
|
cytoglobin
|
|
chr11_+_69832336
|
6.602
|
|
Dlg4
|
discs, large homolog 4 (Drosophila)
|
|
chr2_+_58998938
|
6.590
|
|
Pkp4
|
plakophilin 4
|
|
chrX_-_98615554
|
6.507
|
NM_001177988
NM_019831
|
Zmym3
|
zinc finger, MYM-type 3
|
|
chr6_+_17257720
|
6.489
|
|
Cav1
|
caveolin 1, caveolae protein
|
|
chr19_+_10525829
|
6.484
|
|
|
|
|
chr3_+_117278559
|
6.479
|
|
4833424O15Rik
|
RIKEN cDNA 4833424O15 gene
|
|
chr3_+_117278144
|
6.473
|
NM_029425
|
4833424O15Rik
|
RIKEN cDNA 4833424O15 gene
|
|
chr11_-_5965555
|
6.469
|
|
Camk2b
|
calcium/calmodulin-dependent protein kinase II, beta
|
|
chr19_-_4906607
|
6.454
|
NM_001033128
|
Bbs1
|
Bardet-Biedl syndrome 1 (human)
|
|
chr18_+_37906668
|
6.428
|
|
Pcdhga10
|
protocadherin gamma subfamily A, 10
|
|
chr19_-_7280741
|
6.425
|
NM_134150
|
Otub1
|
OTU domain, ubiquitin aldehyde binding 1
|
|
chr5_-_31504258
|
6.390
|
|
Zfp513
|
zinc finger protein 513
|
|
chr7_+_5031809
|
6.360
|
NM_010147
|
Epn1
|
epsin 1
|
|
chr12_-_73509537
|
6.319
|
|
Rtn1
|
reticulon 1
|
|
chr17_-_24826873
|
6.249
|
NM_011522
|
Syngr3
|
synaptogyrin 3
|
|
chr15_+_78744862
|
6.241
|
|
Pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase
|
|
chr11_+_101017197
|
6.225
|
NM_134028
|
Tubg2
|
tubulin, gamma 2
|
|
chr3_-_152003317
|
6.210
|
NM_001162375
NM_174868
|
Fam73a
|
family with sequence similarity 73, member A
|
|
chr1_+_75542840
|
6.171
|
NM_009208
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3
|
|
chr2_+_58999028
|
6.163
|
|
Pkp4
|
plakophilin 4
|
|
chr12_+_112997057
|
6.159
|
NM_001025360
NM_001081959
NM_008450
|
Klc1
|
kinesin light chain 1
|
|
chr7_+_133968166
|
6.130
|
NM_001146347
|
Fam57b
|
family with sequence similarity 57, member B
|
|
chr3_+_107698740
|
6.118
|
NM_010360
|
Gstm5
|
glutathione S-transferase, mu 5
|
|
chr4_+_43455122
|
6.110
|
NM_011571
|
Tesk1
|
testis specific protein kinase 1
|
|
chr4_-_75779379
|
6.071
|
NM_001014288
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D
|
|
chr16_+_35155621
|
6.052
|
NM_001012765
|
Adcy5
|
adenylate cyclase 5
|
|
chr15_+_81767370
|
6.037
|
|
Csdc2
|
cold shock domain containing C2, RNA binding
|
|
chrX_-_119510890
|
6.024
|
NM_138742
|
Nap1l3
|
nucleosome assembly protein 1-like 3
|
|
chr17_-_46162111
|
6.024
|
NM_001110266
NM_001110267
NM_001110268
|
Vegfa
|
vascular endothelial growth factor A
|
|
chr15_+_103333554
|
5.998
|
NM_008800
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent
|
|
chr11_+_78002309
|
5.998
|
|
Rab34
|
RAB34, member of RAS oncogene family
|
|
chr1_+_74331503
|
5.993
|
NM_001039509
NM_025580
|
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia
|
|
chr8_+_41393330
|
5.991
|
NM_030110
|
Efha2
|
EF-hand domain family, member A2
|
|
chr5_+_37633353
|
5.978
|
|
Crmp1
|
collapsin response mediator protein 1
|
|
chr11_-_116515602
|
5.950
|
NM_030206
|
Cygb
|
cytoglobin
|
|
chr2_+_164793796
|
5.882
|
|
Slc12a5
|
solute carrier family 12, member 5
|
|
chr7_+_4642529
|
5.877
|
NM_001003920
NM_001168572
|
Brsk1
|
BR serine/threonine kinase 1
|
|
chr11_+_102622745
|
5.864
|
NM_001110778
NM_009613
|
Adam11
|
a disintegrin and metallopeptidase domain 11
|
|
chr5_-_31597792
|
5.853
|
NM_022424
|
Fndc4
|
fibronectin type III domain containing 4
|
|
chr19_+_5689759
|
5.823
|
|
Map3k11
|
mitogen-activated protein kinase kinase kinase 11
|
|
chr15_-_58654964
|
5.772
|
NM_175212
|
Tmem65
|
transmembrane protein 65
|
|
chr1_+_60466462
|
5.744
|
NM_001198570
NM_001198571
NM_198127
|
Abi2
|
abl-interactor 2
|
|
chr3_-_57651409
|
5.696
|
|
Pfn2
|
profilin 2
|
|
chrX_+_130829929
|
5.641
|
|
Tmem35
|
transmembrane protein 35
|
|
chr8_+_96334879
|
5.633
|
|
Gnao1
|
guanine nucleotide binding protein, alpha O
|
|
chr11_-_97914582
|
5.624
|
NM_146028
|
Stac2
|
SH3 and cysteine rich domain 2
|
|
chr7_-_31911754
|
5.577
|
|
Scn1b
|
sodium channel, voltage-gated, type I, beta
|
|
chr4_-_57002965
|
5.560
|
|
6430704M03Rik
|
RIKEN cDNA 6430704M03 gene
|
|
chr19_+_53385022
|
5.550
|
|
Mxi1
|
Max interacting protein 1
|
|
chr18_+_38456364
|
5.519
|
NM_001164621
|
Rnf14
|
ring finger protein 14
|
|
chr12_-_73509668
|
5.512
|
|
Rtn1
|
reticulon 1
|
|
chr5_+_34339351
|
5.511
|
|
Nat8l
|
N-acetyltransferase 8-like
|
|
chr4_+_43394824
|
5.486
|
NM_001037709
|
Rusc2
|
RUN and SH3 domain containing 2
|
|
chr19_-_37281951
|
5.482
|
|
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr18_+_37303622
|
5.466
|
NM_001003672
|
Pcdhac2
|
protocadherin alpha subfamily C, 2
|
|
chr4_-_148473897
|
5.420
|
NM_019781
|
Pex14
|
peroxisomal biogenesis factor 14
|
|
chr18_+_37906739
|
5.349
|
NM_033593
|
Pcdhga10
|
protocadherin gamma subfamily A, 10
|
|
chr4_-_133053867
|
5.347
|
|
Trnp1
|
TMF1-regulated nuclear protein 1
|
|
chr11_-_118770859
|
5.345
|
|
Rbfox3
|
RNA binding protein, fox-1 homolog (C. elegans) 3
|
|
chr1_+_60466499
|
5.341
|
|
Abi2
|
abl-interactor 2
|
|
chr19_+_8994860
|
5.332
|
NM_024256
|
B3gat3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I)
|
|
chr6_-_124915441
|
5.324
|
|
Cops7a
|
COP9 (constitutive photomorphogenic) homolog, subunit 7a (Arabidopsis thaliana)
|
|
chr17_-_27957398
|
5.323
|
NM_001033279
NM_001044719
|
D17Wsu92e
|
DNA segment, Chr 17, Wayne State University 92, expressed
|
|
chr4_-_133054355
|
5.321
|
NM_001081156
|
Trnp1
|
TMF1-regulated nuclear protein 1
|
|
chr13_-_49243351
|
5.316
|
NM_029361
|
Wnk2
|
WNK lysine deficient protein kinase 2
|
|
chr4_+_43394949
|
5.306
|
|
Rusc2
|
RUN and SH3 domain containing 2
|
|
chr1_+_60466568
|
5.277
|
|
Abi2
|
abl-interactor 2
|
|
chr5_-_31504605
|
5.259
|
NM_175311
|
Zfp513
|
zinc finger protein 513
|
|
chr12_+_3365102
|
5.233
|
NM_008445
|
Kif3c
|
kinesin family member 3C
|